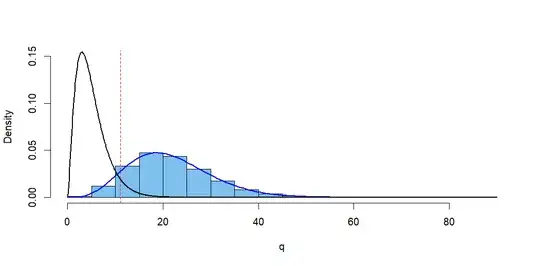
We roll a die $n = 450$ times to assess whether it is fair, The null hypothesis $H_0$ (that each of the six faces has probability $1/6$ of showing at any one roll) is to be tested at the 5% level of significance.
Let $X_i$ be the number of times face $i$ shows in $450$ rolls. We use the test statistic
$$Q = \sum_{i=1}^6 (X_i -75)^2/75 \stackrel{\text{aprx}}{\sim} \mathsf{Chisq}(\text{df} = 5),$$ rejecting $H_0$ for $Q \ge 11.0705.$
In fact, the die is biased so that the probabilities of the respective faces are truly $p_1 = (1/6, 1/6, 1/9, 2/9, 1/6, 1/6),$ not $p_0 = (1/6, 1/6, 1/6, 1/6, 1/6, 1/6).$
What is the power of the test against this alternative? That is, what is the probability that $H_0$ is (correctly) rejected?
Note: This is not (yet) a homework or examination question. I will post/accept my solution if I do not see an Answer with a better one.

sum(c(1/6,1/6,1/9,2/9,1/6,1/6))returns $1.$ Also $1/9 + 2/9 = 1/6 + 1/6 =1/3.$ – BruceET Nov 23 '17 at 07:23