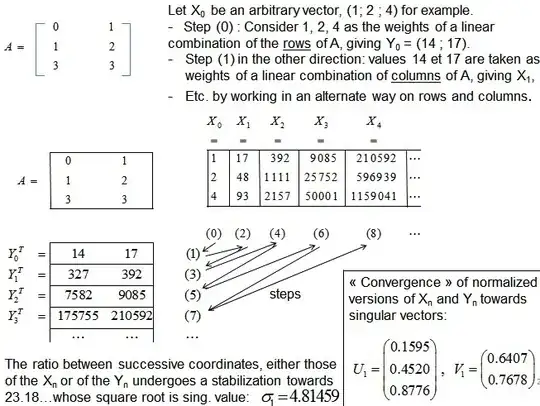
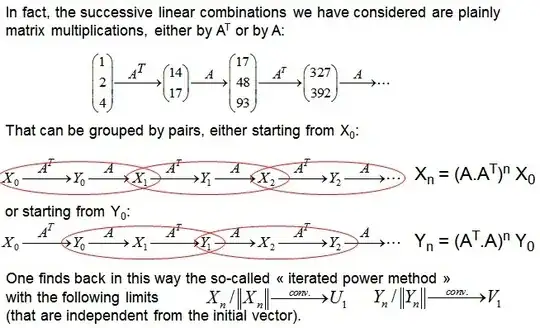
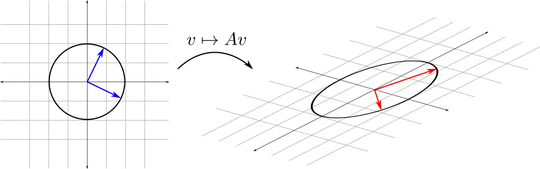
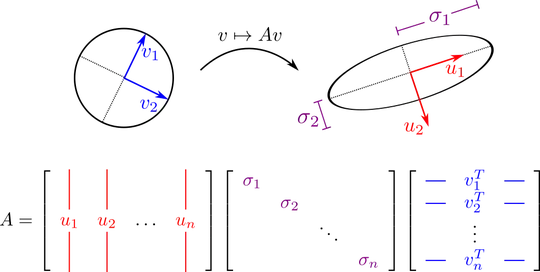
I am not a mathematician, so I need to understand what SVD does and WHY more than how it works exactly from the math perspective. (I understand at least what is the decomposition though).
This guy on youtube gave the only human explanation of SVD saying, that the U matrix maps "user to concept correlation" Sigma matrix defines the strength of each concept, and V maps "movie to concept correlation" given that initial matrix M has users in the rows, and movie (ratings) in the columns.
He also mentioned two concept specifically "sci fi" and "romance" movies. See the picture below.
My questions are:
How SVD knows the number of concepts. He as human mentioned two - sci fi, and romance, but in reality in resulting matrices are 3 concepts. (for example matrix U - that one with blue titles - has 3 columns not 2).
How SVD knows what is the concept after all. I mean, what If i shuffle the columns randomly how SVD then knows what is sci fi, what is romance. I mean, I suppose there is no rule, group the concepts together in the column order. What if scifi movie is the first and last one? and not first 3 columns in the initial matrix M?
What is the practical usage of either U, Sigma or V matrices? (Except that you can multiply them to get the initial matrix M)
Is there also any other possible human explanation of SVD than the guy up provided, or it is the only one possible function? Matrices of correlations.