I've recently coded up a suite of algorithms for computing the persistent homology for various data sets (small data sets roughly around 30 data points). A question has come to my mind about how to find an optimal stopping criterion for generating the Vietoris-Rips sequence.
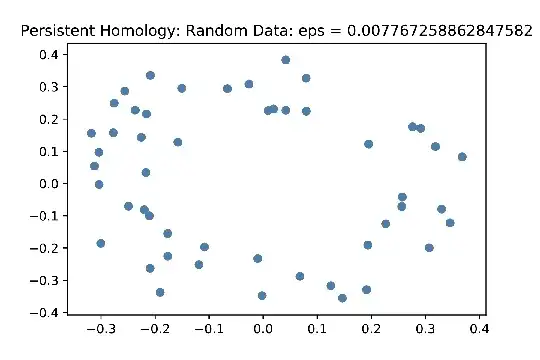
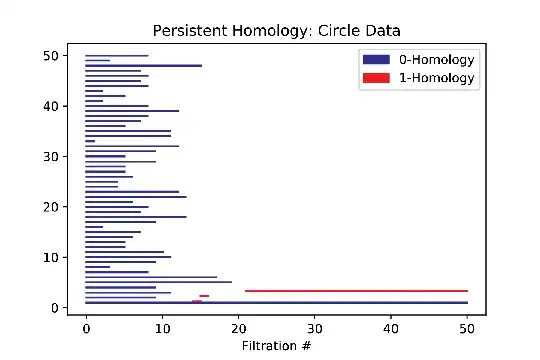
The Vietoris-Rips sequence is constructed by forming simplicial complexes of the data points by systematically increasing the threshold distance allowed to draw an edge between two points. Eventually, as the threshold distance increases, the image is filled with more and more simplexes until eventually the resulting space is a simply connected blob. For those unfamiliar with persistent homology, the desired result of the algorithm is to produce a graph called a bar code which is a visual representation of the homological features that are "born" at some stage of the Vietoris-Rips sequence, and eventually "die" when enough simplexes are drawn in the data. As an example, below are screen images of an initial data set (looking vaguely circular), a later shot after some simplexes have been filled in, and the resulting barcode.
My question is: what is a good stopping criterion for determining the largest distance needed in the Vietoris-Rips sequence to accurately capture all of the interesting (i.e. persistent) topological features of the data set? The problem with these algorithms is that they are extremely expensive, generating enormous matrices (for context, I relied almost exclusively on "A Roadmap for the Computation of Persistent Homology" by Otter et al. for my specific implementation). Two clear ideas for stopping criteria have come to mind:
- Compute the Vietoris-Rips sequence up to some fraction of the maximal distance seen in the data set (essentially guesstimating, unless there are good lower bounds on when to expect topological features to die out).
- Simply stopping when all ${N\choose 2}$ edges are drawn between the data points, most likely generating an unnecessarily long filtration and costing much time and computational energy to generate.
This is my first venture into topological data analysis. It would be great to know what literature there is published on cutting the computational complexity of these algorithms.