I have an imbalanced dataset like so:
df['y'].value_counts(normalize=True) * 100
No 92.769441
Yes 7.230559
Name: y, dtype: float64
The dataset consists of 13194 rows and 37 features.
I have tried numerous attempts to improve the performance of my models by oversampling and undersampling to balance the data, One Class SVM for outlier detection, using different scores, hyperparametre tuning, etc. Some of these methods have improved the performance slightly, but not as much as I would like:
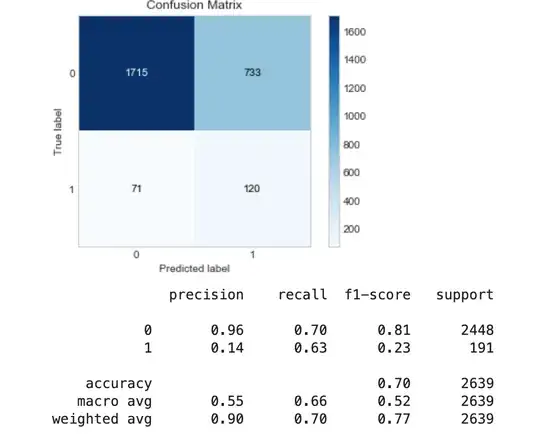
Applying RandomUnderSampling:
from imblearn.under_sampling import RandomUnderSampler
rus = RandomUnderSampler(random_state=42)
X_train_rus, y_train_rus = rus.fit_resample(X_train, y_train)
Define and fit AdaBoost classifier using undersampled data
ada_rus = AdaBoostClassifier(n_estimators=100, random_state=42)
ada_rus.fit(X_train_rus,y_train_rus)
y_pred_rus = ada_rus.predict(X_test)
evaluate_model(y_test, y_pred_rus)
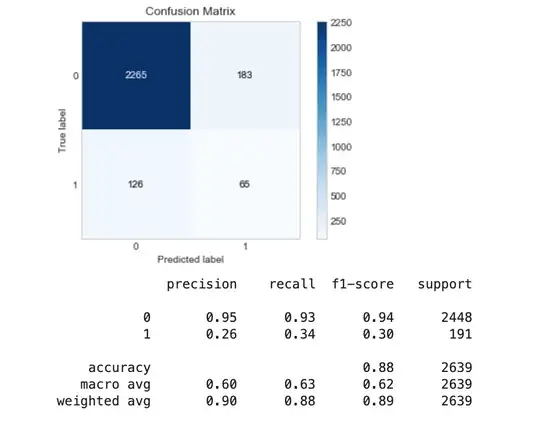
Using Oversampling techniques such as SMOTE:
# SMOTE
from imblearn.over_sampling import SMOTE
upsample minority class using SMOTE
sm = SMOTE(random_state=42)
X_train_sm, y_train_sm = sm.fit_sample(X_train, y_train)
Define and fit AdaBoost classifier using upsample data
ada_sm = AdaBoostClassifier(n_estimators=100, random_state=42)
ada_sm.fit(X_train_sm,y_train_sm)
y_pred_sm = ada_sm.predict(X_test)
compare predicted outcome through AdaBoost upsampled data with real outcome
evaluate_model(y_test, y_pred_sm)
I then decided to attempt removing rows with missing data from samples from the majority class as I saw this in an article. I did this gradually by increasing the threshold (thresh) parameter in pandas dropna function, and each time I removed more rows, the performance improved. Finally, I removed all rows from the majority class with missing data like so:
df_majority_droppedRows = df.query("y == 'No'").dropna()
df_minority = df.query("y == 'Yes'")
dfWithDroppedRows = pd.concat([df_majority_droppedRows, df_minority])
print(dfWithDroppedRows.shape)
(1632, 37)
This reduced the number of rows I have dramatically down to 1632 and changed the distribution in the target variable such that what was perviously the minority class('Yes') was now the majority class:
Yes 58.455882
No 41.544118
Name: y, dtype: float64
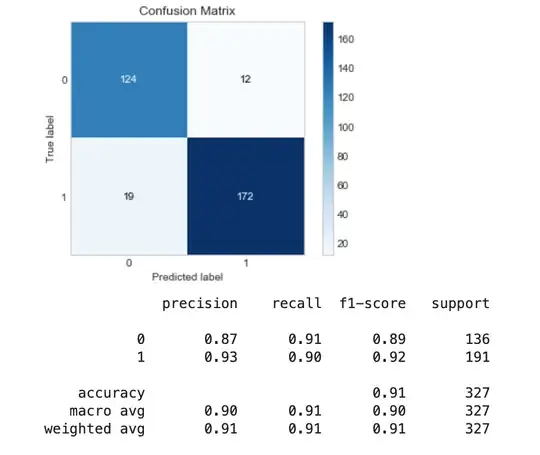
Testing the model, I found it performed best, with high recall and precision values.
So my questions are,
Why did this method outperform other oversampling and undersampling techniques?
Is it acceptable that what was previously a minority class is now the majority class or can this cause overfitting?
Is it realistic to build a model that relies on input with no missing data for the majority class samples?
EDIT
In response to the questions in the comment by @Ben Reiniger:
- I dealt with the missing values like in the data by using KNNImputer for numeric data and SimpleImputer for categorical data like so:
def preprocess (X):
# define categorical and numeric transformers
numeric_transformer = Pipeline(steps=[
('knnImputer', KNNImputer(n_neighbors=2, weights="uniform")),
('scaler', StandardScaler())])
categorical_transformer = Pipeline(steps=[
('imputer', SimpleImputer(strategy='constant', fill_value='missing')),
('onehot', OneHotEncoder(handle_unknown='ignore'))])
preprocessor = ColumnTransformer(transformers=[
('cat', categorical_transformer, selector(dtype_include=['object'])),
('num', numeric_transformer, selector(dtype_include=['float64','int64']))
])
X = pd.DataFrame(preprocessor.fit_transform(X))
return X
- After dropping rows, I defined the feature of matrix and the target, preprocessed and then split the data, like so:
# make feature matrix and target matrix
X = dfWithDroppedRows.drop(columns=['y'])
y = dfWithDroppedRows['y']
encode target variable
from sklearn.preprocessing import LabelEncoder
le = LabelEncoder()
y = le.fit_transform(y)
preprocess feature matrix
X=preprocess(X)
Split data into training and testing data
X_train, X_test, y_train, y_test = train_test_split(X, y, stratify=y, test_size=0.2, random_state=42)
- Finally, to calculate if the missing values are identically distributed (originally) in the two classes, I ran the following
np.count_nonzero(df.query("y == 'No'").isna()) / df.query("y == 'No'").size
0.2791467938526762
np.count_nonzero(df.query("y == 'Yes'").isna()) / df.query("y == 'Yes'").size
0.24488639582979205
So the majority class has about 28% missing data and the minority class has about 25% missing data.