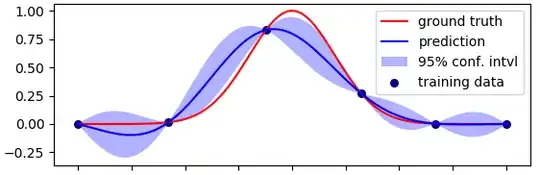
I have an idea but I am not sure if it is correct. Please feel free to express whatever opinion or emotions you might have about the following solution.
Classification and regression tasks are very similar. If done, for example, via neural networks, then a network for regression will differ from the corresponding network for classification only in activation function of the output neuron and the loss function.
The idea is to bin the target variable for the regression task, make a classification on the binned labels, and then use predict_proba to get the probability of the predicted values to be in a certain interval.
The prediction probability for the initial regression task can be estimated based on the results of predict_proba for the corresponding classification.
This is how it can be done for the same toy problem as shown on the picture in the question. The task is to learn a 1-D gaussian function
def gaussian(x, mu, sig):
return np.exp(-np.square((x-mu)/sig)/2)
given some training data.
I build the following neural network in Keras:
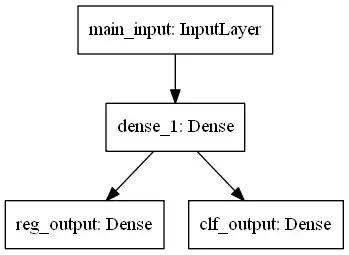
The network is trained simultaneously for both classification and regression. It splits only in the last layer. The input is one-dimensional.
The hidden layer has 10 neurons. The output layer for regression is one neuron with the linear activation. The output layer for classification has several softmax neurons. Their amount depends on how many bins are filled with target variables.
In this toy example, I have 6 training data points:
# training data
x_train = np.atleast_2d([3.2487, -1.2235, -10.0, 10.0, -5.7789, 6.6834]).T
y_train = gaussian(x_train, mu, sig)
I divide the whole range were the target variable changes (0 to 1) into 10 bins. Each bin is 0.1 wide. The amount of bins can be thought of as a hyper-parameter. The more bins the closer the classification problem to the corresponding regression problem. But too many bins is probably not good.
# Binning the target variable
hist, bin_edges = np.histogram(y_train, bins=np.linspace(0, 1, 11))
y_c = np.digitize(y_train, bin_edges)
n_classes = len(np.unique(y_c))
# Binarize targets for classification
lb = LabelBinarizer()
y_b = lb.fit_transform(y_c)
The training data fall into three bins. You can see in the picture why. The four points far on each side (two on the left side, and two on the right side) are all in one bin, and each of the remaining points in the middle is in a separate bin. The remaining seven bins are empty. So, the output layer for the classification task has 3 softmax neurons. I use 1-hot encoding for the labels.
This is the network:
# NNet with one input and two outputs: one for reg, another for clf
main_input = Input(shape=(1,), dtype='float32', name='main_input')
hidden = Dense(10, input_dim=1, activation='tanh')(main_input)
reg_output = Dense(1, activation='linear', name='reg_output')(hidden)
clf_output = Dense(n_classes, activation='softmax', name='clf_output')(hidden)
model = Model(inputs=[main_input], outputs=[reg_output, clf_output])
Different loss functions are used for classification and regression. I also assign different loss weights which can be thought of as another hyper-parameter.
model.compile(optimizer='adam',
loss={'reg_output': 'mse', 'clf_output': 'categorical_crossentropy'},
loss_weights={'reg_output': 1., 'clf_output': 0.2})
Training:
model.fit({'main_input': x_train},
{'reg_output': y_train, 'clf_output': y_b},
epochs=1000, verbose=0)
Running model.predict gives prediction for the regression and prediction probabilities for classification.
# Prediction for both classification and regression
y_pred, pred_proba_c = model.predict({'main_input': x})
Each row of the array pred_proba_c contains probabilities of putting a test point to one of three classes. I estimate a regression's analogue of predict_proba by taking the maximum of these three probabilities.
# This is a regression's analogue of predict_proba
r_pred_proba = np.max(pred_proba_c, axis=1)
This is the result. The prediction probability is shown in the bottom half of the picture.
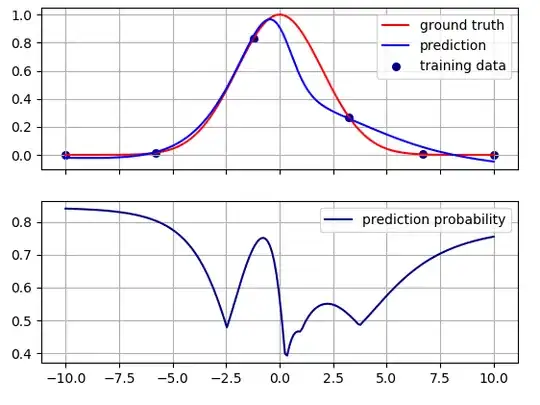
Intuitively, the probability is high where there are training data, and it decreases in the regions between the training data. The model becomes less sure about its predictions far from the training data.
The maxima of the prediction probability are not exactly at the training points. This might be because there is no exact correspondence between the underlying classification and regression problems. They are related but they are not the same, and the relationship between them depends on the values of the hyper-parameters, and the learning algorithm. For example, if I change the loss weights,
model.compile(optimizer='adam',
loss={'reg_output': 'mse', 'clf_output': 'categorical_crossentropy'},
loss_weights={'reg_output': 1., 'clf_output': 1})
I get the following picture:

Now the prediction probability values are different but the qualitative behavior is the same.
The complete code is as follows:
import numpy as np
import matplotlib.pyplot as plt
from keras.models import Model
from keras.layers import Input, Dense
from sklearn.preprocessing import LabelBinarizer
np.random.seed(1)
x = np.atleast_2d(np.linspace(-10, 10, 200)).T
mu = 0
sig = 2
def gaussian(x, mu, sig):
return np.exp(-np.square((x-mu)/sig)/2)
# training data
x_train = np.atleast_2d([3.2487, -1.2235, -10.0, 10.0, -5.7789, 6.6834]).T
y_train = gaussian(x_train, mu, sig)
# Binning the target variable
hist, bin_edges = np.histogram(y_train, bins=np.linspace(0, 1, 11))
y_c = np.digitize(y_train, bin_edges)
n_classes = len(np.unique(y_c))
# Binarize targets for classification
lb = LabelBinarizer()
y_b = lb.fit_transform(y_c)
# NNet with one input and two outputs: one for reg, another for clf
main_input = Input(shape=(1,), dtype='float32', name='main_input')
hidden = Dense(10, input_dim=1, activation='tanh')(main_input)
reg_output = Dense(1, activation='linear', name='reg_output')(hidden)
clf_output = Dense(n_classes, activation='softmax', name='clf_output')(hidden)
model = Model(inputs=[main_input], outputs=[reg_output, clf_output])
model.compile(optimizer='adam',
loss={'reg_output': 'mse', 'clf_output': 'categorical_crossentropy'},
loss_weights={'reg_output': 1., 'clf_output': 0.2})
model.fit({'main_input': x_train},
{'reg_output': y_train, 'clf_output': y_b},
epochs=1000, verbose=0)
# Prediction for both classification and regression
y_pred, pred_proba_c = model.predict({'main_input': x})
# This is a regression's analogue of predict_proba
r_pred_proba = np.max(pred_proba_c, axis=1)
f, ax = plt.subplots(2, sharex=True)
ax[0].plot(x, gaussian(x, mu, sig), color="red", label="ground truth")
ax[0].scatter(x_train, y_train, color='navy', s=30, marker='o', label="training data")
ax[0].plot(x, y_pred, 'b-', color="blue", label="prediction")
ax[0].legend(loc='best')
ax[0].grid()
ax[1].plot(x, r_pred_proba, color="navy", label="prediction probability")
ax[1].legend(loc='best')
ax[1].grid()
plt.show()