When using an XGB model in the context of binary classification, I observed that the test estimates given by predict_proba were close but not equal to the results I obtained by summing the outputs of the corresponding leaves for each observation and then converting that sum (which is a log-odds) to probability using the logistic function $\frac{e^x}{1+e^x}$. Why does this happen? What am I not taking into account and how can I recreate the results of predict_proba with the individual outputs from the trees?
My code (based on the example given in the XGB docs page) starts by fitting the model:
from sklearn.datasets import load_iris
from xgboost import XGBClassifier
from sklearn.model_selection import train_test_split
data = load_iris()
X_train, X_test, y_train, y_test = train_test_split(data['data'][data['target']!=2],
data['target'][data['target']!=2], test_size=.2, random_state=42)
# create model instance
bst = XGBClassifier(n_estimators=5, max_depth=2, learning_rate=0.5,
objective='binary:logistic', colsample_bytree=0.3)
# fit model
bst.fit(X_train, y_train)
After doing this I was able to retreive the individual trees of the model and hard-code them into a function to obtain the tree outputs in an array:
def empiric_model(X):
# t0,...,t4 is for the trees and f0, f1, f2 are the features
t0 = -0.952112317 if (X['f2']<3).all() else 0.869410515
t1 = -0.462368667 if (X['f0']<5.5).all() else 0.51116854
t2 = -0.571811914 if (X['f2']<3).all() else 0.569128871
t3 = 0.403415382 if (X['f1']<3).all() else 0.0914318636 if ((3<=X['f1'])&(X['f1']<3.0999999)).all() else -0.11495477 if ((3.0999999<=X['f1'])&(X['f1']<3.4000001)).all() else -0.417713255
t4 = 0.318558067 if (X['f1']<3).all() else 0.0555250905 if ((3<=X['f1'])&(X['f1']<3.20000005)).all() else -0.309806734
return np.array([t0, t1, t2, t3, t4])
Then I compared both bst.predict_proba(X_test)[:,1] and the sum of the leaves outputs converted into probability. The results of that comparison are shown in the next graph:
# sum of leaf-outputs for each test observation
empirics = np.array([empiric_model(pd.DataFrame(X_test[i:i+1,:], columns=['f0','f1','f2','f3'])).sum() for i in np.arange(X_test.shape[0])])
# convert that into proba using the logistic function
empiric_probas = np.exp(empirics)/(1+np.exp(empirics))
predicted_probas = bst.predict_proba(X_test)[:,1]
plt.scatter(x=np.arange(empiric_probas.size), y=empiric_probas, c='b', label='empiric')
plt.scatter(x=np.arange(empiric_probas.size), y=predicted_probas, c='r', label='predicted')
plt.xticks(np.arange(empirico_probas.size))
plt.yticks(np.arange(0,1.1,0.2))
plt.legend(loc='center right')
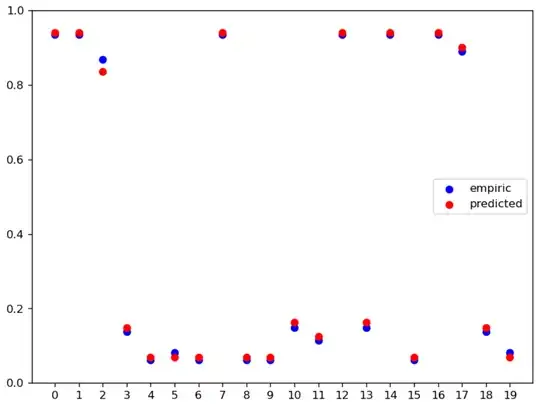 I'll explicitly show these values:
I'll explicitly show these values:
empiric_probas is:
array([0.9353348 , 0.9353348 , 0.86878725, 0.13712985, 0.06216319,
0.08233362, 0.06216319, 0.9353348 , 0.06216319, 0.06216319,
0.14927792, 0.11448531, 0.9353348 , 0.14927792, 0.9353348 ,
0.06216319, 0.9353348 , 0.8905786 , 0.13712985, 0.08233362])
while predicted_probas is:
array([0.9411262 , 0.9411262 , 0.8354762 , 0.1493973 , 0.06825472,
0.06825472, 0.06825472, 0.9411262 , 0.06825472, 0.06825472,
0.16242756, 0.12502052, 0.9411262 , 0.16242756, 0.9411262 ,
0.06825472, 0.9411262 , 0.89994967, 0.1493973 , 0.06825472])
Which as you can see are similar but not equal.