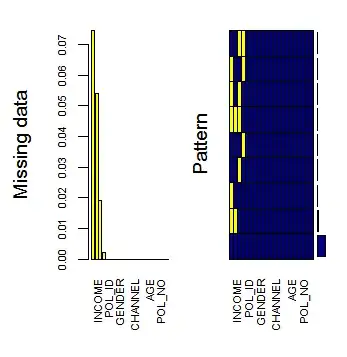
I have a large data-frame (155257 x 21 to be specific) with only a few missing values. Say, some 2.16% of the values need to be imputed. The values are floating point numbers.
I'd like to use a method that is much faster than it is accurate, because of the size of the data-set and the fact that I don't have much to lose in a speed-accuracy tradeoff.
Running missForest() takes several hours while Hmisc's impute() function gives unsatisfactory results.
What functions in R might be useful in such (or similar) case?