my goal is to make prediction (good or bad data) on sensor data. I tried a lot, but failed to shape my data to get the desired output.
scenario:
I have multiple timestamped (time as it self is not important) measurements files with each containing 1700 datapoints of 2 features. The first column is the timestamp (in ns). t0 = 1 ns after the start of the meassurement. The other two columns are the data points at that timestamp namely the depth and intensity.
Here is a header of such a file:
columns = ['timestamp', 'Intensity', 'depth']
array([[ 1. , 79. , -0.5273184 ],
[ 14. , 94. , -0.56211778],
[ 29. , 102. , -0.59692583],
[ 43. , 109. , -0.57392274],
[ 57. , 111. , -0.55091889]])
[1700 rows (timestamps) and 2 features])
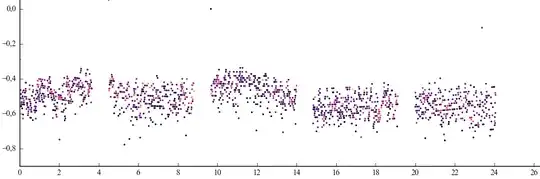
A good measurement looks like that (image at the end of the post):
The x-axis represent the time-axis calulated to a length in mm. The y-axis is the depth-axis and the color represent the intensity of that datapoint.
What i have done:
I created a list with all my files
file_names_list = [file_name for file_name in os.listdir(path_for_csv_data) if file_name.endswith('.txt')]and looped thru that listIn each interation of that loop i created a DataFrame and reshaped it and appeded to data_list:
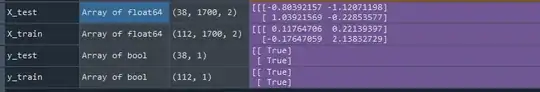
data_list = [] for single_file_name in file_names_list: #create df pandas_data_frame = pd.read_csv(os.path.join(path_for_csv_data, single_file_name), index_col=0, header=0, decimal = '.', delimiter = ';')#drop the timestamp because i dont need it pandas_data_frame.drop(columns = ['timestamp'], axis = 1, inplace = True) #reshape each file to (1700, 2) pandas_data_frame.values.reshape(-1, len(pandas_data_frame.columns)) #should be (1700, 2) #append all files to a list, to have all the data in one place data_list.append(pandas_data_frame) #shape is now (150, 1700, 2)I splited my data into train and test data (used RobustScaler on train data), loaded my labels and created a sequential model:
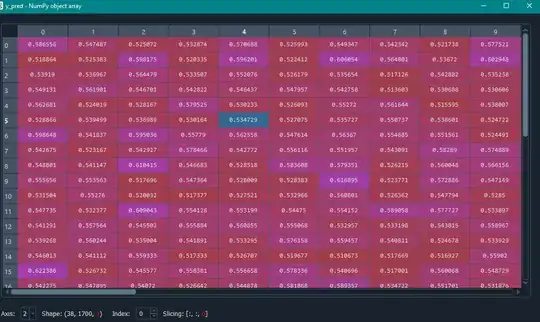
model = Sequential() model.add(Dense(100, input_shape = (1700,2), activation = 'relu')) model.add(Dropout(0.2)) model.add(Dense(40, activation = 'relu')) model.add(Dense(1, activation = 'sigmoid')) model.compile(optimizer = 'adam', loss = 'mse', metrics = ['accuracy'])Then a fited my data into my model but if i do so, i get a prediction of every datapoint (look image at the end of this post) in a shape of (x, 1700, 1), not on a file itself (shape of (x, 1)).
model.fit(X_train, y_train) y_pred = model.predict(X_test)
My Questions:
- What to do, to get the right output?
- Is my way of prepering data right for such a problem? (timestamped data, NOT time series data, like predicting bitcoin price)) -> i just want to feed my model a numpy array with 1700 rows and 2 columns und want a output wether the data is good or bad.