Unsupervised k-NN
Unlike k-means, the unsupervised k-nn does not associate a label to instances. All it can do is tell you what instances in your training data is k-nearest to the point you are polling for.
For example:
import numpy as np
from sklearn.neighbors import NearestNeighbors
samples = [[0, 0, 2], [1, 0, 0], [0, 0, 1]]
neigh = NearestNeighbors(2, 0.4)
neigh.fit(samples)
neigh.kneighbors([[0, 0, 1.3]], 2, return_distance=False)
array([[2, 0]]...)
You can see that this returned the index of the k-nearest points, and not the label.
k-means
This algorithm is completely different. The k here denotes the number of assumed classes that exist in your dataset. For example if you have unlabeled pictures of red and green apples, you know that $k=2$. The algorithm will then move the centroids (the average of the cluster distributions) to a stable solution.
Here is an example:
Let's first make some artificial Gaussian distributed data.
import numpy as np
import matplotlib.pyplot as plt
params = [[[ 0,1], [ 0,1]],
[[ 5,1], [ 5,1]],
[[-2,5], [ 2,5]],
[[ 2,1], [ 2,1]],
[[-5,1], [-5,1]]]
n = 300
dims = len(params[0])
data = []
y = []
for ix, i in enumerate(params):
inst = np.random.randn(n, dims)
for dim in range(dims):
inst[:,dim] = params[ix][dim][0]+params[ix][dim][1]*inst[:,dim]
label = ix + np.zeros(n)
if len(data) == 0: data = inst
else: data = np.append( data, inst, axis= 0)
if len(y) == 0: y = label
else: y = np.append(y, label)
num_clusters = len(params)
print(y.shape)
print(data.shape)
(1500,)
(1500, 2)
plt.scatter(data[:,0], data[:,1])
plt.show()

The k-means algorithm from scratch
class Kmeans(object):
def __init__(self, k=1):
self.k = k
def train(self, data, verbose=1):
shape = data.shape
ranges = np.zeros((shape[1], 2))
centroids = np.zeros((shape[1], 2))
for dim in range(shape[1]):
ranges[dim, 0] = np.min(data[:,dim])
ranges[dim, 1] = np.max(data[:,dim])
if verbose == 1:
print('Ranges: ')
print(ranges)
centroids = np.zeros((self.k, shape[1]))
for i in range(self.k):
for dim in range(shape[1]):
centroids[i, dim] = np.random.uniform(ranges[dim, 0], ranges[dim, 1], 1)
if verbose == 1:
print('Centroids: ')
print(centroids)
plt.scatter(data[:,0], data[:,1])
plt.scatter(centroids[:,0], centroids[:,1], c = 'r')
plt.show()
count = 0
while count < 100:
count += 1
if verbose == 1:
print('-----------------------------------------------')
print('Iteration: ', count)
distances = np.zeros((shape[0],self.k))
for ix, i in enumerate(data):
for ic, c in enumerate(centroids):
distances[ix, ic] = np.sqrt(np.sum((i-c)**2))
labels = np.argmin(distances, axis = 1)
new_centroids = np.zeros((self.k, shape[1]))
for centroid in range(self.k):
temp = data[labels == centroid]
if len(temp) == 0:
return 0
for dim in range(shape[1]):
new_centroids[centroid, dim] = np.mean(temp[:,dim])
if verbose == 1:
plt.scatter(data[:,0], data[:,1], c = labels)
plt.scatter(new_centroids[:,0], new_centroids[:,1], c = 'r')
plt.show()
if np.linalg.norm(new_centroids - centroids) < np.finfo(float).eps:
print("DONE!")
break
centroids = new_centroids
self.centroids = centroids
self.labels = labels
if verbose == 1:
print(labels)
print(centroids)
return 1
def getAverageDistance(self, data):
dists = np.zeros((len(self.centroids),))
for ix, centroid in enumerate(self.centroids):
temp = data[self.labels == ix]
dist = 0
for i in temp:
dist += np.linalg.norm(i - centroid)
dists[ix] = dist/len(temp)
return dists
def getLabels(self):
return self.labels
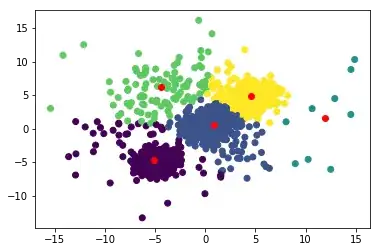
And the results
kmeans = Kmeans(5)
kmeans.train(data)
Ranges:
[[-15.42553872 14.88894099]
[-13.33192554 16.15415347]]
Centroids:
[[-11.39200726 -10.71208054]
[ 3.73634888 -8.9230959 ]
[ 6.17589734 -10.66376228]
[ 0.78973744 -0.44245535]
[
9.29524253 9.59127574]]
Initialize, the red points are the random centroids
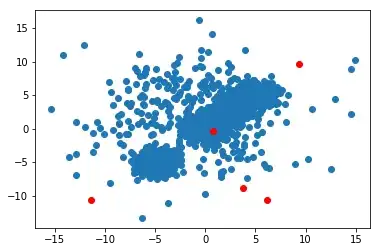
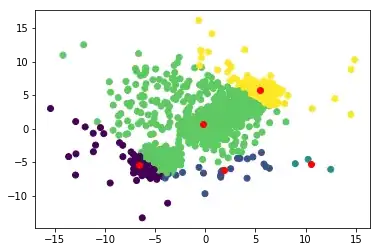
Iteration 1
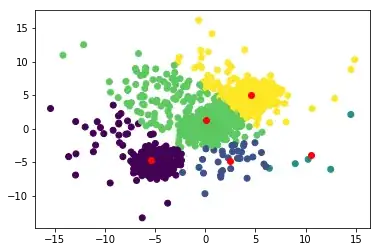
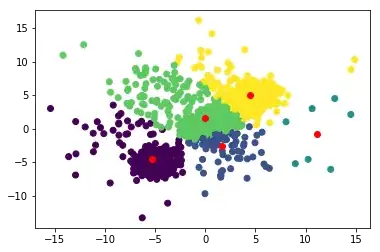
and after a few iterations