import pandas as pd
import numpy as np
from sklearn import set_config
set_config(transform_output = "pandas")
np.random.seed(0) #reproducible results
df = pd.read_csv('Dados ML.xlsx - Literature dataset.csv', header=1)
#Remove the DOI column
df = df.drop(columns='DOI')
#Drop amino column
amino_col = 'Dominant amino acid group of enzymes'
df = df.drop(columns=amino_col)
#Replace commas with a dot and
and '-' with NaN to allow numerical conversion
df = df.apply(lambda col: col.str.replace(',', '.').replace('-', np.nan), axis=1)
#Identify categorical vs numeric columns
ligand_col = 'Ligand precursor functional group'
categorical_cols = [ligand_col,]
numeric_cols = [col for col in df.columns if col not in categorical_cols]
#Convert numbers (currently strings) to numeric types
df[numeric_cols] = df[numeric_cols].astype(np.float64)
#Drop unavailable targets
target_col = 'Reusability: Relative activity after 7 reuse cycles (%)'
df = df.drop(index=df.index[ df[target_col].isna() ])
df = df.loc[df[target_col] != 0] #Drop zeros assuming they're invalid
def quick_hist(df, title=''):
df_num = df.select_dtypes(np.float64)
axs = df_num.hist(bins=50, grid=False, figsize=(7, 5))
for i, name in enumerate(df_num.columns):
col_num = df.columns.to_list().index(name)
axs.flatten()[i].set_title(f'[{col_num}] ' + name[:min(16, len(name))], fontsize=9)
plt.suptitle(title, fontsize=12)
plt.tight_layout()
plt.show()
#Features/target for preprocessing
features = df.drop(columns=target_col)
target = df[target_col]
quick_hist(features, 'features before outlier removal')
outliers_bool = (
(features.iloc[:, 2] > 10) |
(features.iloc[:, 3] > 50) |
(features.iloc[:, 5] > 50) |
(features.iloc[:, 10] > 200_000) |
(features.iloc[:, 11] > 100_000)
)
print('Detected', outliers_bool.sum(), 'outliers')
df_inliers = df.drop(index=features.index[ outliers_bool ])
features_inliers = df_inliers.drop(columns=target_col)
target_inliers = df_inliers[target_col]
quick_hist(features_inliers, 'features after outlier removal')
drop_outliers = False
if drop_outliers:
print('Dropping outliers'.upper())
use_features = features_inliers
use_target = target_inliers
else:
print('NOT dropping outliers')
use_features = features
use_target = target
#Split off a test test to be used as a final unbiased measure
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
use_features, use_target, test_size=0.15, stratify=pd.cut(use_target, bins=5),
shuffle=True, random_state=1000
)
#Use imputation to fill in missing data
from sklearn.impute import SimpleImputer
from sklearn.compose import ColumnTransformer
cat_transformer = SimpleImputer(strategy='most_frequent')
num_transformer = SimpleImputer(strategy='median')
numeric_features = [col for col in numeric_cols if col != target_col]
imputer = ColumnTransformer(
[
('Categorical imputer', cat_transformer, categorical_cols),
('Numeric imputer', num_transformer, numeric_features)
],
verbose_feature_names_out=False
)
#Encode categorical column
from sklearn.preprocessing import OneHotEncoder
ligand_encoder = OneHotEncoder(sparse_output=False, drop='if_binary')
cat_encoder = ColumnTransformer(
[
('Ligand encoder', ligand_encoder, [ligand_col]),
],
remainder='passthrough',
verbose_feature_names_out=False
)
#Compose a preprocessor pipeline
from sklearn.pipeline import make_pipeline
preprocessor = make_pipeline(imputer, cat_encoder)
#Try a random forest
from sklearn.ensemble import RandomForestRegressor
from sklearn.model_selection import cross_val_score, KFold, LeaveOneOut
#Shuffle the data per fold for kfold
kfold_cv = KFold(n_splits=6, shuffle=True, random_state=0)
loo_cv = LeaveOneOut()
model = RandomForestRegressor(random_state=np.random.RandomState(0))
#Dummy regressor
from sklearn.dummy import DummyRegressor
model = DummyRegressor(strategy='median')
#Another model I tried
from sklearn import ensemble
model = ensemble.GradientBoostingRegressor(learning_rate=0.1/4, random_state=np.random.RandomState(0))
#Compose pipeline with model
pipeline = make_pipeline(preprocessor, model)
scores = cross_val_score(
estimator=pipeline,
X=X_train, y=y_train,
cv=kfold_cv, scoring='r2', n_jobs=-1
)
print('median score:', np.median(scores).round(3))
print('scores:', scores.round(3))
"""
median score: 0.569
scores: [-0.008 0.473 0.613 0.539 0.799 0.599]
"""
Let's inspect each fold
from matplotlib import pyplot as plt
from sklearn.model_selection import cross_validate
from sklearn.metrics import mean_squared_error, r2_score
#Get complete results
cv_results = cross_validate(
estimator=pipeline,
X=X_train, y=y_train,
cv=kfold_cv, scoring='r2', n_jobs=-1,
return_estimator=True,
return_indices=True
)
trn_predictions = []
val_predictions = []
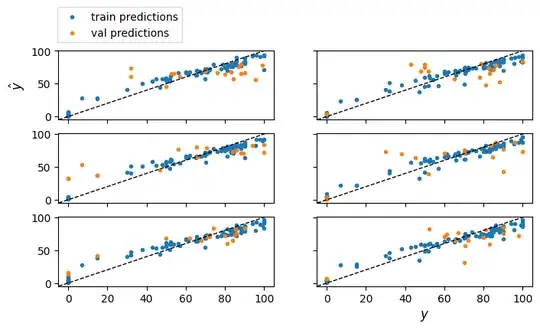
f, axs = plt.subplots(3, 2, figsize=(8, 4), sharex=True, sharey=True)
axs = axs.flatten()
for fold in range(kfold_cv.get_n_splits()):
fold_estimator = cv_results['estimator'][fold]
fold_trn_indices = cv_results['indices']['train'][fold]
fold_val_indices = cv_results['indices']['test'][fold]
trn_predictions.append(
fold_estimator.predict(X_train.iloc[fold_trn_indices])
)
val_predictions.append(
fold_estimator.predict(X_train.iloc[fold_val_indices])
)
#Print fold's RMSE and R2
fold_val_y = y_train.iloc[fold_val_indices]
print(f'Fold {fold}\n val RMSE:',
mean_squared_error(fold_val_y, val_predictions[fold], squared=False))
print(' val R2:',
r2_score(fold_val_y, val_predictions[fold]), '\n')
#Scatter of train and val predictions for fold
fold_trn_y = y_train.iloc[fold_trn_indices]
axs[fold].scatter(fold_trn_y, trn_predictions[fold], marker='.', label='train predictions')
axs[fold].scatter(fold_val_y, val_predictions[fold], marker='.', label='val predictions')
axs[fold].axline(xy1=(0, 0), slope=1, color='black', linewidth=1, linestyle='--')
axs[-1].set_xlabel('$y$', fontsize=12)
axs[0].set_ylabel('$\hat{y}$', fontsize=12)
axs[0].legend(bbox_to_anchor=(0.6, 1.7))
plt.show()
#Use leave-one-out CV
from sklearn.model_selection import cross_validate
cv_results = cross_validate(
estimator=pipeline,
X=X_train,
y=y_train,
cv=loo_cv,
scoring='neg_mean_absolute_error',
return_estimator=True,
return_indices=True,
n_jobs=-1
)
#Get out-of-sample predictions
predictions = np.empty(y_train.shape)
for fold in range(loo_cv.get_n_splits(X_train)):
predictions[fold] = cv_results['estimator'][fold].predict(
X_train.iloc[cv_results['indices']['test'][fold]]
).item()
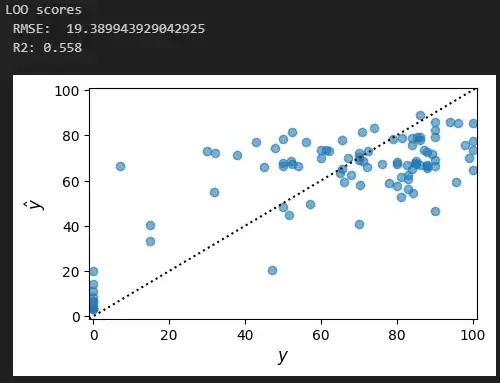
print('LOO scores')
print(' RMSE: ', mean_squared_error(y_train, predictions, squared=False))
print(' R2:', r2_score(y_train, predictions).round(3))
val_indices_flat = np.concatenate(cv_results['indices']['test'])
from matplotlib import pyplot as plt
plt.scatter(y_train.iloc[val_indices_flat], predictions, alpha=0.6)
plt.axline(xy1=(0, 0), slope=1, color='black', linestyle=':')
plt.xlim(-1, 101)
plt.ylim(-1, 101)
plt.xlabel('$y$', fontsize=12)
plt.ylabel('$\hat{y}$', fontsize=12)
plt.gcf().set_size_inches(5, 3)
"""
Final evaluation on the test set
fitted_pipeline = pipeline.fit(X_train, y_train)
test_predictions = pipeline.predict(X_test)
test_r2 = r2_score(y_test, test_predictions)
test_rmse = mean_squared_error(y_test, test_predictions, squared=False)
print('Final test set scores')
print('RMSE', round(test_rmse, 2))
print('R2:', round(test_r2, 3))
"""