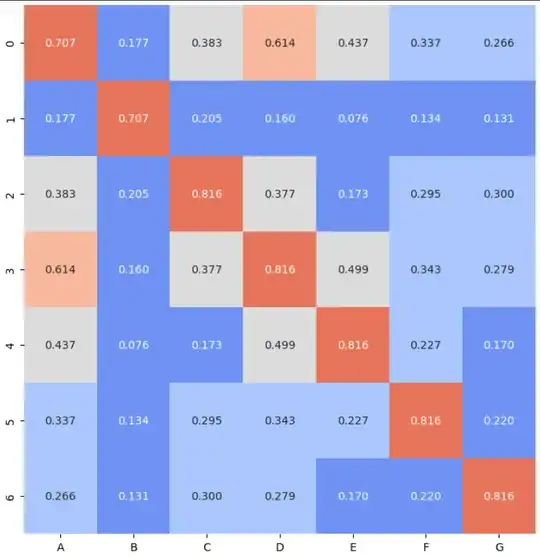
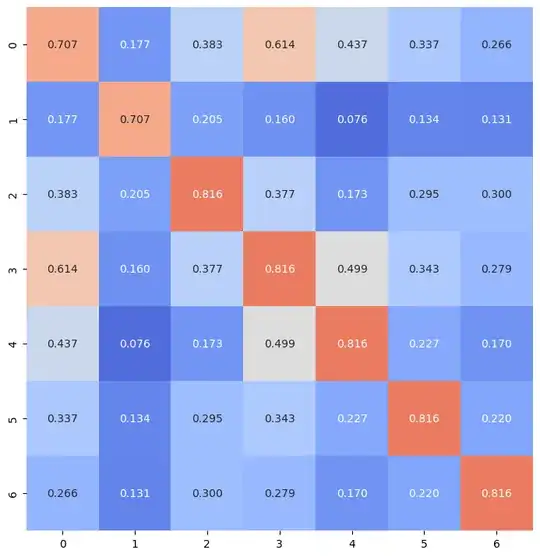
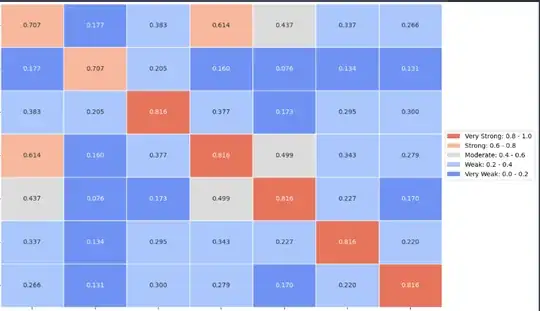
For the following dataframe of a chi2 correlation study, i started to plot a heatmap:
import pandas as pd
import numpy as np
columns = ['A', 'B', 'C', 'D', 'E', 'F', 'G']
results = np.array([[0.70709269, 0.17683162, 0.38328705, 0.61449242, 0.43709035, 0.33675627, 0.2661715 ],
[0.17683162, 0.70709268, 0.20520211, 0.16044232, 0.07607822, 0.13364355, 0.13093324],
[0.38328705, 0.20520211, 0.81649658, 0.37683897, 0.17308779, 0.29541159, 0.29975079],
[0.61449242, 0.16044232, 0.37683897, 0.81649658, 0.4991043 , 0.34257853, 0.2786975 ],
[0.43709035, 0.07607822, 0.17308779, 0.4991043 , 0.81649658, 0.22700152, 0.17041603],
[0.33675627, 0.13364355, 0.29541159, 0.34257853, 0.22700152, 0.81649658, 0.22018705],
[0.2661715 , 0.13093324, 0.29975079, 0.2786975 , 0.17041603, 0.22018705, 0.81649658]])
df_matrix = pd.DataFrame(results, columns=columns)
category_bounds = [0, 0.2, 0.4, 0.6, 0.8, 1.0]
categories = ['Very Weak', 'Weak', 'Moderate', 'Strong', 'Very Strong']
df_heatmap = pd.DataFrame(df_matrix, index=df_matrix.index, columns=df_matrix.columns)
colors = sns.color_palette('coolwarm', len(categories))
cmap = ListedColormap(colors)
fig, ax = plt.subplots(figsize=(10, 8))
sns.heatmap(df_heatmap, annot=True, cmap=cmap, fmt=".3f", cbar=False, ax=ax, linecolor='white')
plt.subplots_adjust(left=0.25, top=0.95)
plt.show()
But, for some reason (I suppose it is due to rounding the values), 0.71 and 0.82 are plotting in the same color. Can someone give me some guidance on what the problem is?