I have a medium size data set (7K) of patient age, sex, and pre-existing conditions. Age of course is from 0-101, sex is 1 for male, 2 for female, and -1 for diverse. All the pre-conditions are Boolean. The outcome, death is also Boolean.
Regardless of how I scale the data (I tried lots of scalers), I always get a warning:
FitFailedWarning: Estimator fit failed. The score on this train-test partition for
these parameters will be set to nan.
This traces back to: ValueError("Unknown label type: %r" % y_type)
ValueError: Unknown label type: 'unknown'
If I take out the age and sex columns, the error goes away. There are definitely no text, missing, or weird values here.
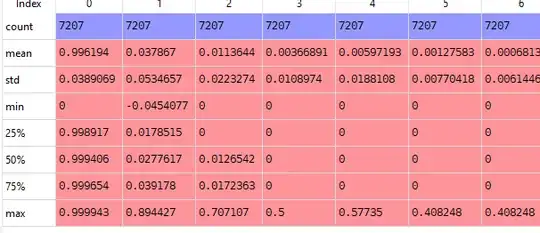
If I look at my rescaled data, it looks as I would expect it to look.
If I drastically simplify the data, it works.
import numpy as np
import pandas as pd
from sklearn import preprocessing
array = np.array([[42, 1, False, False, False, False, False, False, False, False, False, False, False],
[72, 1, False, False, True, False, False, False, False, False, False, True, False],
[77, 2, False, False, False, False, False, False, False, False, True, True, False],
[36, 1, False, False, False, False, False, False, False, False, False, False, False],
[42, 1, False, False, False, False, False, False, False, False, True, False, False],
[82, 1, False, False, False, True, False, False, False, False, False, True, False],
[71, 2, False, False, False, False, False, False, False, False, False, True, False],
[36, -1, False, False, False, False, False, False, False, False, True, False, False],
[52, 1, False, False, False, False, False, False, False, False, False, False, False],
[52, 1, False, False, False, False, False, False, False, True, False, True, True],
[77, 2, False, False, False, False, False, False, True, False, True, True, False],
[46, 1, False, False, False, False, False, False, False, False, False, False, False],
[45, 1, False, False, False, False, False, False, False, False, False, False, False],
[88, 1, False, False, False, False, False, True, False, False, False, True, True],
[79, 2, False, True, True, False, False, False, False, False, False, True, True],
[36, -1, True, False, False, False, False, False, False, False, False, False, False]])
X = array[:,0:12]
Y = array[:,12]
scaler = preprocessing.MinMaxScaler().fit(X)
rescaledX = scaler.transform(X)
from sklearn.model_selection import KFold
from sklearn.model_selection import cross_val_score
from sklearn.linear_model import LogisticRegression
kfold = KFold(n_splits=3, shuffle=True, random_state=7) # split the data into training and test sets for k-fold validation
model = LogisticRegression(solver='lbfgs') # set up model of a linear regression
results = cross_val_score(model, rescaledX, Y, cv=kfold)
print("Accuracy: %.3f%% (%.3f%%)" % (results.mean()100.0, results.std()100.0))
It would be awesome if someone has an idea of what might be wrong, or how to troubleshoot further.